1. Introduction
Annokey is a command line tool for annotating gene lists with the results of a key-term search of the NCBI Gene database and linked PubMed article abstracts. Its purpose is to help users prioritise genes by relevance to a domain of interest, such as "breast cancer" or "DNA repair" etcetera. The user steers the search by specifying a ranked list of keywords and terms that are likely to be highly correlated with their domain of interest.
2. License
Annokey is open source software, released under the 3-clause BSD license.
3. Quick start guide
- Install Annokey on your computer; see Section 4 below.
- Prepare your two input files: 1) a genes file in CSV format and 2) a key-terms file in text format. See Section 6 for more details about the inputs.
- Choose between online and offline mode. See Section 7 for more detail about the differences.
- Online mode is suitable for a relatively short number of genes (< 100).
- Offline mode is suitable for relatively large number of genes (>= 100) or in cases where you plan to do lots of searches.
Quick start online mode
- Run annokey on the inputs and save the output (replace
user@example.comwith your real email address):
annokey --genes genes.csv --terms terms.txt --online --email user@example.com > genes_out.csv
Quick start offline mode
- Populate the gene cache from the NCBI snapshot. See section 9 for how to do this.
- Run annokey on the inputs and save the output:
annokey --genes genes.csv --terms terms.txt > genes_out.csv
Quick start PubMed
Annokey can optionally search for the key-terms in the PubMed article titles and abstracts which are linked from the records in the NCBI Gene database. PubMed search can be used with both online and offline modes of Annokey. Note that PubMed search can significantly increase the search time, so it is not turned on by default. PubMed search may require Annokey to access data online which requires you to supply an email address.
Here is the online example command from above extended with PubMed search:
annokey --genes genes.csv --terms terms.txt --pubmed --online --email user@example.com > genes_out.csv
Here is the offline example command from above extended with PubMed search (note the email address is still required):
annokey --genes genes.csv --terms terms.txt --pubmed --email user@example.com > genes_out.csv
4. Installation
Annokey currently requires version 2.7 of Python.
The best way to install Annokey is to use the following command:
pip install git+https://github.com/bjpop/annokey.git
This will automatically download and install the dependencies of Annokey.
5. Features
Annokey's main features are:
Online and offline search
- Online search of the NCBI databases directly over a network connection. Pros: you get the latest up-to-date information. Cons: retrieving lots of data over the network can be relatively slow.
- Offline search utilises Annokey's local cache of the NCBI databases. Pros: searching the local cache is much faster than online search and does not need a network connection. Cons: the cache data may be incomplete or out of date.
The NCBI provides an entire copy of the gene database in XML format at ftp.ncbi.nlm.nih.gov. Annokey can use this XML file to automatically populate its local cache. We also provide a tool called get_ncbi_gene_snapshot_xml.py (described in Section 9 below) for automatically downloading this XML copy of the database.
Flexible search terms
Search terms can be specified as literal terms or as regular expressions.
- Literal terms are easy to use but an exact match must be found in the database. Sometimes exact match is too restrictive, for example, issues with upper- and lower-case letters.
- Regular expressions are much more flexible than literal terms, but they require more expertise from the user.
Search results reported in summary and detailed form
The search results are provided in summary form, as extra annotations to the input gene list, and also in a more detailed report, as a HTML document. An example report is shown in Section 6.
- Summary annotations provide a quick overview of the relevance of a key phrase to a gene, and can be incorporated into a workflow using spreadsheets.
- The detailed report provides more information than the summary, and contains hyperlinks back to the online NCBI databases for easy reference.
6. Inputs and outputs
Annokey's main inputs are:
- A CSV file representing a set of genes, one gene per row. Rows may contain other arbitrary columns, but one column must contain the gene name.
- A list of search terms sorted in descending order of priority.
Annokey's main outputs are:
- A CSV file representing an annotated version of the input CSV file. Each row of the CSV file (representing a given gene) is given three extra columns summarising the search results. The extra columns are explained below in an example.
- A HTML report file showing more detailed search results for each gene. This includes hyperlinks back to the relevant entries in the NCBI website for easy follow-up investigation, plus detailed lists containing the context of each key-term match in each field.
Simple example of inputs and outputs
For example, if your input gene CSV file contains the following rows (this is an intentionally simple example, real data will often contain more columns on each row):
Gene
XRCC2
PALB2
BRCA1
and your input search term file contains the following lines:
DNA repair
breast cancer
the output CSV file produced by Annokey will look like this:
Gene,Highest Rank,Highest Rank Term,Total Matched Entries
XRCC2,1,DNA repair,11
PALB2,1,DNA repair,48
BRCA1,1,DNA repair,408
Consider the first case, for XRCC2:
XRCC2,1,DNA repair,11
In addition to the gene name, the output row contains three extra columns:
- Highest Rank: the rank of the highest-ranked search term which was found in the search. In this case the rank is 1, which corresponds to the rank of the search term "DNA repair". Search terms are ranked according to their order in the search terms file. The term on the first line is highest ranked (most important), followed by the second line, and so on.
- Highest Rank Term: this is the search-term corresponding to the highest ranked match. In the above example the highest ranked matching term is "DNA repair".
- Total Matched Entries: the sum of the number of database fields matched for every search term. For each search term we count the number of database fields where the term is matched, and then sum them for all terms. If two different terms match in the same field we still count them separately. This number gives you a crude measure of how closely related a given gene is to the entire collection of search terms.
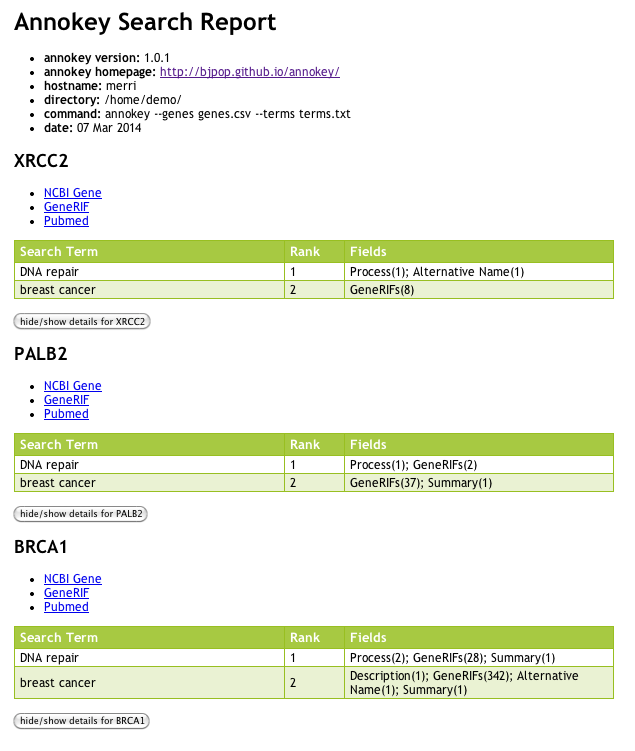
Below is a screen shot of the search report generated for the example above. You can see that the report is much more detailed than the CSV annotations. The report is divided into genes. Each gene report contains a table showing all the matches for each search term. The rank of each matching search term is shown in the table, along with a list of database fields where the term matched, and the corresponding frequency of matches in those fields are shown in brackets.
Input formatting requirements
The input CSV file must be formatted as follows:
- The file may be comma or tab separated. Annokey looks at the filename extension to guess what format is used (".csv" for comma separated files, ".tsv" for tab separated files) but you may override this with the
--delimiter {comma|tab}command line argument. - The first row in the file must be a header row.
- Each row may have one or more columns. One of the columns must contain the gene name, and the header for that row must be "Gene". Annokey assumes that you will use the standard gene names that appear in the Gene database (NCBI gene database nomenclature.
Here is an example valid gene file in comma separated format:
Sample,Study,Gene,Variant
s100,XYZ,BRCA1,snp
s100,XYZ,XRCC2,indel
s888,ABC,PALB2,snp
s100,ABC,CHECK2,snp
Notice that there are multiple columns in each row. Annokey only requires the "Gene" column, the other columns are ignored as far as the search is concerned, but are preserved in the output file.
The search terms file must be formatted as follows:
- Each search term must appear on a separate line.
- A search term is a regular expression, using the Python regular expression syntax. The Python regular expression HOWTO provides a good tutorial on the topic.
- The simplest kind of regular expression is just a collection of one or more literal words, such as "breast cancer". This kind of term will match exactly the text in the term and nothing else. For example "DNA" will not match "dna" because they are not an exact match.
- More complex regular expressions can be used to make matching more flexible. For example "[Bb]reast [Cc]ancer" will match "Breast Cancer", "Breast cancer", "breast Cancer" and "breast cancer".
- Search terms are ranked based on their relative order in the file. The search term on line 1 has rank 1 and so on. Thus the most important search term appears on the first line of the file. The second most important term appears on the second line, and so forth.
A literal search term might look like this:
breast cancer
A regular expression search term might look like this:
[Bb]reast [Cc]ancer(s)?
This regular expression is more flexible because it will match with upper and lower case characters at the start of each word, and it also allows for an optional "s" at the end of the second word.
7. Online versus offline search
Annokey obtains its search results by querying the NCBI gene database and the linked PubMed article abstracts. It can access the data in two ways:
- By sending queries to the online database via the Entrez programming interface.
- By searching in a locally cached copy of the data.
Online queries give you access to the most up-to-date version of the database, but at the cost of being much slower, and requiring an internet connection. The offline version searches in a local copy of the database stored on your computer. Offline search is potentially much faster than online search and does not need an internet access. However, your local copy of the database may not necessarily contain the most up-to-date data.
Annokey treats the Gene cache and the PubMed cache differently for these reasons:
- PubMed is much larger than the Gene database. It is easy to download an entire copy of the Gene database, but less so for PubMed. Also PubMed has stricter licensing conditions than Gene.
- Existing PubMed database entries are unlikely to be modified, the opposite is true for existing Gene database entries.
For these reasons Annokey allows you to populate a local cache with a copy of the entire Gene database for a given organism, but it does not allow you to do the same with PubMed. If you specify the --pubmed command line argument, Annokey will collect all the PubMed references made from each gene entry, and fetch all (and only) those PubMed articles directly from NCBI. However, it will keep a local cache of any previously requested PubMed articles, and fetch them from the cache if they are ever requested again in the future. This avoids the time delay introduced by the network request, and reduces the load on the PubMed server. We recommend that you do not use the PubMed search on large numbers of genes (>=100) because the cost of requesting large numbers of PubMed articles can greatly slow down the search process.
By default Annokey keeps its cache of the NCBI Gene Database in a directory called genecache, and a cache of the PubMed article summaries in pubmedcache, however you can specify an alternative locations via the --genecache DIR and --pubmedcache DIR command line arguments. The structure of the cache directories is discussed in the technical details section below.
8. How to use Annokey from the command line
You can get help about the Annokey command line by using annokey -h (or --help), which will produce this output:
usage: annokey [-h] [--version] [--online] [--cachesnapshot FILE]
[--organism ORGANISM] [--email EMAIL_ADDRESS] [--genecache DIR]
[--pubmedcache DIR] [--terms FILE] [--genes FILE]
[--log FILENAME] [--delimiter {comma,tab}] [--report FILENAME]
[--pubmed]
Search NCBI for genes of interest, based on concept-keyword search.
optional arguments:
-h, --help show this help message and exit
--version show program's version number and exit
--online Search gene information from online (NCBI).
--cachesnapshot FILE Populate the gene cache from downloaded XML snapshot
of NCBI gene database
--organism ORGANISM Name of the organism to search
--email EMAIL_ADDRESS
Your email address. This is required by NCBI for
online queries. You do not need to supply an email
address for cached queries.
--genecache DIR Save a cache of the downloaded results from NCBI gene
into this directory
--pubmedcache DIR Save a cache of the downloaded results from NCBI
pubmed into this directory
--terms FILE The tab separated file containing the search terms to
be searched.
--genes FILE The tab separated file containing the gene information
including name of the gene, one gene name per line.
--log FILENAME log progress in FILENAME, defaults to annokey_log.txt
--delimiter {comma,tab}
Delimiter for gene file.
--report FILENAME Save a detailed search report as HTML page, defaults
to annokey_report.html
--pubmed Search titles and abstracts in Pubmed entries referred
to in each gene entry
Required arguments
Annokey requires two arguments --terms and --genes, which define the search terms and gene list respectively. The remaining arguments are optional.
Annokey's default mode is to use local offline search in a cached copy of the NCBI Gene. For example, if the file terms.txt contains a list of search terms and the file genes.csv contains the gene information, then the following command is sufficient to perform a search, assuming you have already populated the local cache (see Section 9 for how to do this).
annokey --terms terms.txt --genes genes.csv
Saving the annotated output to a file
Annokey produces a new gene file on standard output. You can save this output to a file using the Unix shell redirection operator ">":
annokey --terms terms.txt --genes genes.csv > annotated_genes.csv
Search report file
Annokey saves a detailed search report in a HTML file. By default this file is called annokey_report.html but you can specify another name using the --report argument:
annokey --report myreport.html --terms terms.txt --genes genes.csv
Log file
As a side-effect, Annokey will produce a log file which contains useful information about the progress of the search. For instance, the log file will record the names of any genes that it could not find in the local cache. By default the log file is written to the file annokey_log.txt, but you can override that with the --log argument:
annokey --log mylogfile.txt --terms terms.txt --genes genes.csv
The log file also records the command line that was used to execute the program. This can come in handy if you want to run the same command again, but you forgot what the arguments were.
Online search
The effectiveness of offline search depends greatly on the status of your local cache of data.
Alternatively, Annokey can be made to perform an online search with the --online argument. This argument also requires you to specify an email address (as per requirements of the NCBI Gene database):
annokey --online --email user@example.com --terms terms.txt --genes genes.csv
Obviously you should use a real email address instead of user@example.com.
Organism
By default Annokey uses the human version of the NCBI Gene database, but you can specify an alternative organism with the --organism argument:
annokey --organism mouse --terms terms.txt --genes genes.csv
Gene file delimiter
Multiple columns in the genes input file can be separated by either commas (so-called CSV format) or tabs (so-called TSV format). If the filename ends in ".csv" then Annokey will assume that the file is in CSV format. If the filename ends in ".tsv" then Annokey will assume that the file is in TSV format. However, you can override this behaviour by specifying the desired format using the --delimiter command line argument. The value of --delimiter can be either comma or tab, for example:
annokey --delimiter comma --terms terms.txt --genes genes.txt
or
annokey --delimiter tab --terms terms.txt --genes genes.txt
PubMed article search
Each Gene database entry contains zero or more links to related PubMed articles which relate to the given gene. Annokey will search for key terms in the linked Pubmed article titles and summaries if you specify the --pubmed flag:
annokey --pubmed --email user@example.com --terms terms.txt --genes genes.txt
Annokey may need to request PubMed articles from the online database, so you must also specify your email address whenever you use the --pubmed flag. Obviously you should use your real email address instead of the example user@example.com.
To reduce the number of network requests to the PubMed online databse Annokey caches each PubMed entry. Future searches of the same article will find it in the cache. This improves the speed of the search and also reduces network traffic.
If Annokey cannot find a particular PubMed entry it will make a note in the logfile and continue searching. Therefore it is a good idea to check the logfile after each search to see whether anything was missed.
9. NCBI Gene snapshot
The following section explains how to create a local cache of the NCBI gene database for a particular organism using the database snapshots.
Summary of steps.
The following two steps summarise how to download the snapshot and use it to populate a local cache. The example illustrates the default behaviour, which is to download the snapshot of the human database. Instructions for other organisms are given below.
- Download and unpack the latest snapshot. This step requires the command
linux.gene2xmlto be in your PATH, or you can specify a path to the command using the--gene2xmlpathoption):
get_ncbi_gene_snapshot_xml.py
The above command will download a copy of the snapshot in ags.gz format. It will convert that into an XML file, whose name will look something like Homo_sapiens_N.xml, where N is an encoding of the snapshot date, for instance Homo_sapiens_20140314054717.xml. The XML file should be several gigabytes in size (it was 12 gigabytes on 14 March 2014).
- Populate the local cache. This step will take a few minutes.
annokey --cachesnapshot Homo_sapiens_20140116041343.xml
This will create a directory called genecache under which the database entries are stored. See Section 10 for more details about the layout of the cache.
After these two steps you are ready to use Annokey with the local cache.
Note carefully Annokey does not check before unpacking the snapshot into the cache directory. If you have a previous cache in the same place it will overwrite the old version without warning. If you want to keep different versions of the cache then you must rename the old cache appropriately. We recommend using a name which contains the date of the snapshot.
More details:
The NCBI provides a complete snapshot of the gene database which is updated regularly. Annokey can use this snapshot to automatically populate its local cache of the data. In many cases this is the most efficient way to use Annokey.
In the example above the snapshot is stored in the file called Homo_sapiens_20140116041343.xml. It stores a record for each gene in the human database. Annokey will process the XML file and populate the local gene database cache. Future searches can just use the cache directly, and will be substantially faster.
Unfortunately there is no corresponding snapshot of the NCBI article summary database.
Snapshots of the NCBI gene database for different organisms are provided on the NCBI ftp site. The snapshot is stored in a compressed binary format which can be converted to XML using a tool called gene2xml.
If you want to get a snapshot for another organism you must specify its FTP path with the --filepath command line argument. Currently all organisms are stored underneath /gene/DATA/ASN_BINARY/ on the NCBI ftp site. For example, the following two commands download and unpack the data for mouse:
Download the mouse snapshot and covert to XML:
get_ncbi_gene_snapshot_xml.py --filepath /gene/DATA/ASN_BINARY/Mammalia/Mus_musculus.ags.gz
Unpack the mouse XML snapshot into its own local cache:
annokey --organism mouse --cachesnapshot Mus_musculus_20140314054725.xml
Note that you must specify the --organism for non-human data (otherwise Annokey will assume it is human). You can name the organism anything you like ("mouse" instead of "Mus musculous"), as long as you remember to use the same name when you run a search command.
The get_ncbi_gene_snapshot_xml.py program supports several command line arguments. You can view help information by running get_ncbi_gene_snapshot_xml.py -h:
usage: get_ncbi_gene_snapshot_xml.py [-h] [--verbose VERBOSE]
[--ftpsite FTPSITE]
[--filepath GENEFILEPATH] [--force]
[--downloadonly] [--convertonly GENEFILE]
[--gene2xmlpath PROGRAMPATH]
This program downloads gene data from the ftp server and converts data to
xml file by using linux.gene2xml tool. This program checks the last modified
time of the database in the server, when it tries to download the gene
database from the server. If the same version of database is in the local
directory already, this program does not download the database from server. If
you want to download the database anyway, please use the --force option. If
you already have the database file that you need to look up and want to
convert the file, please use the --convertonly option.
optional arguments:
-h, --help show this help message and exit
--verbose VERBOSE Prints progress messages. The users have to pass the
level of messages. 0: No progress messages. 1: Prints
what stage the program is on. 2: Prints detail
information on progress. 3: Prints callstack when
error occurs.
--ftpsite FTPSITE The ftp site to be connected for downloading gene
database. Default is NCBI server
(ftp.ncbi.nlm.nih.gov).
--filepath GENEFILEPATH
The full file path in the ftp site that would be
downloaded. The path should start from the root of the
ftp site. For example, if you want to download gene
database of Archaea from NCBI FTP site, you should
pass
/gene/DATA/ASN_BINARY/Archaea_Bacteria/Archaea.ags.gz
as an argument value. Default is Homo sapiens
(/gene/DATA/ASN_BINARY/Mammalia/Homo_sapiens.ags.gz)
--force Downloads the latest version of data from the server
without regarding the existing file on directory. If
there is the same version in local directory, the file
would be overwritten.
--downloadonly Downloads the latest version of data from the server
but not converts the file to xml.
--convertonly GENEFILE
Converts the input file to xml.
--gene2xmlpath PROGRAMPATH
The path where the gene2xml is in. If the path for
linux.gene2xml is not in your PATH, you should pass
this argument. For example, if linux.gene2xml is in
/usr/local/gene2xml/1.3/bin and not in PATH , then you
should pass either
/usr/local/gene2xml/1.3/bin/linux.gene2xml or
/usr/local/gene2xml/1.3/bin.
10. Technical details
Which parts of NCBI gene and PubMed does Annokey search?
Annokey searches in the following sections of the NCBI gene database. The diagram below illustrates the parts of the NCBI Gene XML schema which are searched by Annokey. Comments in parenthesis provide additional remarks about how the data is processed and, where relevant, provides the "field names" that Annokey associates with the data:
Entrezgene
Entrezgene_track-info
Gene-track/Gene-track_status (we skip entries which are discontinued)
Entrezgene_gene
Gene-ref
Gene-ref_locus (Gene Name)
Gene-ref_desc (Description)
Gene-ref_syn
Gene-ref_syn_E (Synonyms)
Entrezgene_summary (Summary)
Entrezgene_comments
Gene-commentary
Gene-commentary_type (type of commentary)
Gene-commentary_heading (heading of commentary)
Gene-commentary_label (label of commentary)
Gene-commentary_text (GeneRIFs, if type == generif)
Gene-commentary_refs
Pub_pmid/PubMedId (PmIds)
Gene-commentary_products
Gene-commentary_comment
Gene-commentary_text (Pathways, if heading == Pathways)
Gene-commentary_comment
(Interactions, if heading == Interactions)
(Conserved Domains, if heading == Conserved Domains)
(Function, if label == Function)
(Component, if label == Component)
(Process, if label == Process)
NCBI Reference Sequences (RefSeq) (more commentaries)
Entrezgene_properties
Gene-commentary/Gene-commentary_comment
Entrezgene_prot
Prot-ref/Prot-ref_name
Prot-ref_name_E (Alternative Name)
Having collected all the PubMed IDs from a gene entry (as above) Annokey then searches in the corresponding PubMed database entry. The diagram below illustrates the parts of the PubMed XML schema which are searched by Annokey. The parts marked with an asterisk indicate the fields of text where Annokey searches for key phrases.
PubmedArticle
MedlineCitation
Article
JournalTitle (*)
ArticleTitle (*)
Abstract
AbstractText (*)
The structure of the gene cache directory
The local cache of NCBI Gene database entries is structured as follows:
genecache
organism (e.g. human, mouse)
hash_dir (a numbered directory in the range [0,255])
gene_name (e.g. PALB2)
gene_id (XML file, e.g 79728)
At the top level is the genecache directory. This has sub-directories for each organism. This has up to 256 sub-directories numbered 0 to 255. The purpose of these sub-directories is to spread out the stored gene information, so that we don't put all the gene files in one sub-directory. Some computer file systems exhibit poor performance when you put too many files in one directory.
The database records for each gene are stored underneath one of the 256 possible hash directories. The exact hash directory is determined by the formula:
md5(gene_name) % 256
where gene_name is the (string) gene name (such as "PALB2"), md5 is a hash function and % is the modulus operator. We are only using md5 as a way to evenly distribute gene names to directories, so we don't care about its cryptographic properties (particularly its known weaknesses).
For example, the entry for PALB2 is calculated as follows:
md5("PALB2") % 256 = 163
Which means that PALB2 will be found in genecache/human/163/PALB2/79728. The actual data is stored in the file called 79728, which is in XML format. The number 79728 is the NCBI Gene database ID of PALB2, which is also reflected in the URL to access the entry in the online database http://www.ncbi.nlm.nih.gov/gene/79728.
Also note that we normalise the gene name when it is stored in the cache to be all uppercase characters, which means we can reliably find the gene regardless of how it was written in the input gene file or stored in the NCBI database.
The structure of the PubMed cache directory
The local cache of PubMed database entries is structured as follows:
pubmedcache
hash_dir (a numbered directory in the range [0,255])
PubMed ID (XML file, an integer)
Each PubMed entry has a unique integer ID. Similar to the gene cache, we spread all entries across 256 sub-directories, using the formula:
md5(PudMed_ID) % 256
Each PubMed entry is stored in an XML file whose name is equal to its PubMed ID.