Gurita: a command line data analytics and plotting tool
Gurita is a command line tool for transforming, analysing, and visualising tabular data in CSV or TSV format.
At its core Gurita provides a suite of commands, each of which carries out a common data analytics or plotting task. Additionally, Gurita allows commands to be chained together into flexible analysis pipelines.
It is designed to be fast and convenient, and is particularly suited to data exploration tasks. Input files with large numbers of rows (> millions) are readily supported.
Gurita commands are highly customisable, however sensible defaults are applied. Therefore simple tasks are easy to express and complex tasks are possible.
Gurita is implemented in Python and makes extensive use of the Pandas, Seaborn, and Scikit-learn libraries for data processing and plot generation.
Simple example
The following examples use the iris.csv dataset as input. The file contains 150 data rows (plus one heading row) and 5 columns. A pretty display of the first and last 5 rows of the data can be viewed using the following command:
cat iris.csv | gurita pretty
The command generates the following output that is displayed on the terminal:
sepal_length sepal_width petal_length petal_width species
5.1 3.5 1.4 0.2 setosa
4.9 3.0 1.4 0.2 setosa
4.7 3.2 1.3 0.2 setosa
4.6 3.1 1.5 0.2 setosa
5.0 3.6 1.4 0.2 setosa
... ... ... ... ...
6.7 3.0 5.2 2.3 virginica
6.3 2.5 5.0 1.9 virginica
6.5 3.0 5.2 2.0 virginica
6.2 3.4 5.4 2.3 virginica
5.9 3.0 5.1 1.8 virginica
[150 rows x 5 columns]
The following command generates a box plot from iris.csv, such that the Y-axis
represents the sepal_length numerical column, and the X-axis is grouped by the species categorical column.
The goal of this plot is to show the distribution of sepal length of the three different species of iris
flowers contained in the data set.
cat iris.csv | gurita box -x species -y sepal_length
The above command generates an output file called box.species.sepal_length.png
containing the following figure:
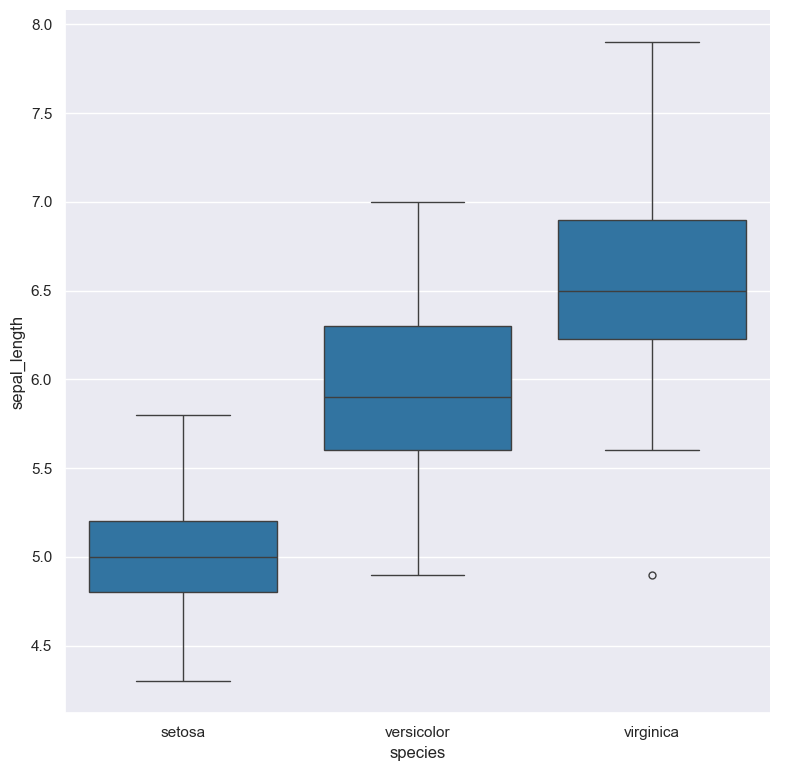
If we wanted to see the median value of sepal_length for each species we can do this with the
following groupby command:
cat iris.csv | gurita groupby --key species --val sepal_length --fun median
species,sepal_length_median
setosa,5.0
versicolor,5.9
virginica,6.5
You can see that the corresponding median values match up with the values shown in the box plot.
Advanced example
The next example illustrates Gurita’s ability to chain commands together:
cat iris.csv | gurita filter 'species != "virginica"' \
+ sample 0.9 \
+ pca \
+ scatter -x pc1 -y pc2 --hue species
The above command has been written on mulitple lines for clarity. Note that the backslash \ indicates a line
break in the command line syntax.
Equivalently, the same command can be written in a single line, like so (where backslashes are no longer required):
cat iris.csv | gurita filter 'species != "virginica"' + sample 0.9 + pca + scatter -x pc1 -y pc2 --hue species
In the command line syntax supported by Gurita, the plus sign + separates each command in a chain. This notation
is fairly novel to Gurita and not something commonly seen in other command line programs.
Input is read from the file called iris.csv on standard input and data is passed from left to right in the chain.
Commands can modify the data as it is passed along.
In this example there are four commands that are executed in the following order:
The
filter 'species != "virginica"'command selects all rows wherespeciesis not equal tovirginica.The filtered rows are then passed to the
sample 0.9command which randomly selects 90% of the remaining rows.The sampled rows are then passed to the
pcacommand which performs principal component analysis (PCA), yielding two extra columns in the data calledpc1andpc2.Finally the pca-transformed data is passed to the
scatter -x pc1 -y pc2 --hue speciescommand which generates a scatter plot ofpc1andpc2(the first two principal components), and colours the points in the plot based on the categoricalspeciescolumn.
The output of this entire Gurita command is the scatter plot below that is saved in a file called scatter.pc1.pc2.species.png
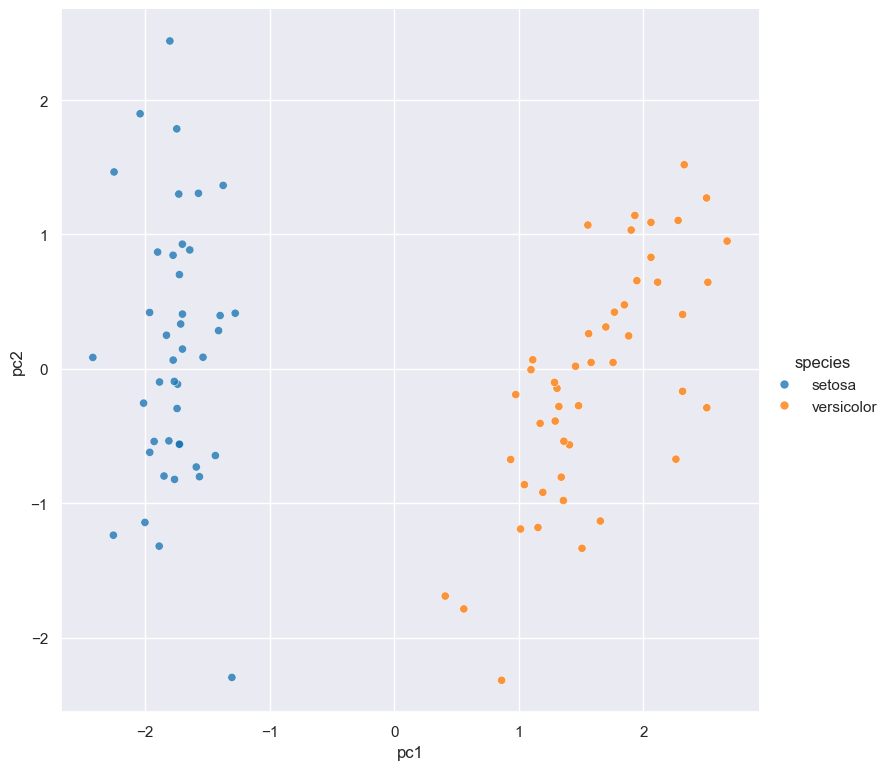
An interesting aspect of this example is that additional columns are added to the data in the pca step, and these extra columns
are then used to define the axes of the following scatter plot.
Command chaining allows complex transformations to be combined together and makes Gurita a very powerful tool.